How can long non-coding RNAs regulate circadian rhythms without producing a protein!?
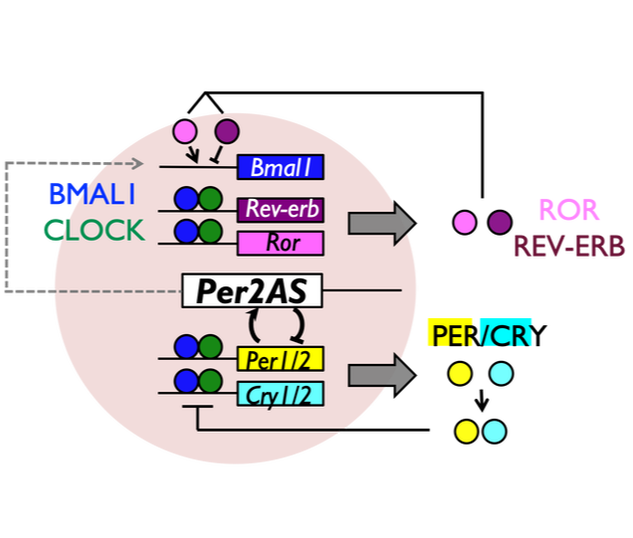
We recently identified a novel non-coding transcript, Per2AS, that appears to play an important role in the mammalian circadian clock system. Interestingly, both of our mathematical and experimental analyses demonstrated that Per2AS confers robustness to the system, and regulates the amplitude of circadian rhythms. How does Per2AS regulate robustness and amplitude without producing a protein? What is the function of Per2AS in vivo? Does Per2AS have additional functions other than regulating circadian rhythms?
What makes circadian rhythms in each organ different from each other?
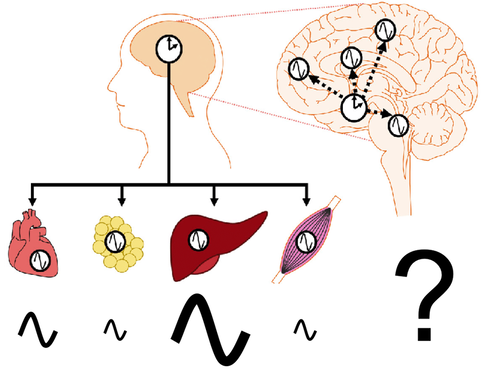
Cell-autonomous circadian clocks drive thousands of rhythmic output genes that, ultimately, produce daily rhythms of many types of physiology and behavior. Interestingly, the number of cycling transcripts is vastly different among mouse tissues, despite the core molecular machinery (i.e., transcription-translation feedback loops) being nearly identical in all the tissues.
We are interested in understanding why some tissues are more robust and produce more cycling genes than others. To answer this, we bioinformatically characterized circadian transcriptome datasets in 12 mouse tissues and found that Rorc, one of the core clock genes, may be the key player to determine which tissues are more robust. We are currently using various experimental tools to validate our bioinformatical prediction and to understand how Rorc contributes to the robustness of the tissues.
Is RNA degradation important for high amplitude circadian gene expression?
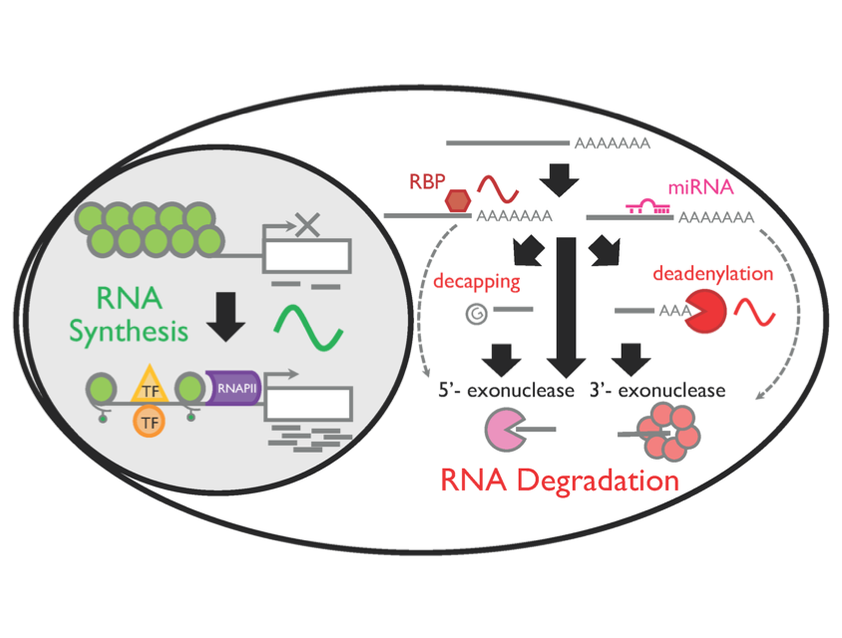
Circadian RNA expression is essential to ultimately regulate a plethora of downstream rhythmic biochemical, physiological, and behavioral processes. Both transcriptional and posttranscriptional mechanisms are considered important to drive rhythmic RNA expression; however, the extent to which each regulatory process contributes to the rhythmic RNA expression remains controversial. To systematically address this, we monitored RNA dynamics using metabolic RNA labeling technology during a circadian cycle in mouse fibroblasts. We find that rhythmic RNA synthesis is the primary contributor of 24-h RNA rhythms, while rhythmic degradation is more important for 12-h RNA rhythms. These rhythms were predominantly regulated by Bmal1 and/or the core clock mechanism, and the interplay between rhythmic synthesis and degradation has a significant impact in shaping rhythmic RNA expression patterns. Interestingly, core clock RNAs are regulated by multiple rhythmic processes and have the highest amplitude of synthesis and degradation, presumably critical to sustain robust rhythmicity of cell-autonomous circadian rhythms. Our study yields invaluable insights into the temporal dynamics of both 24- and 12-h RNA rhythms in mouse fibroblasts.